Research Project - Details
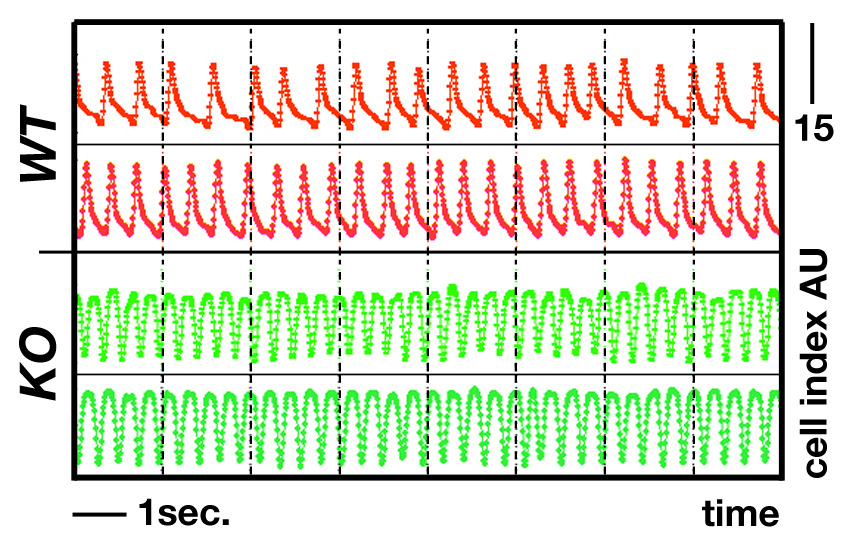
Analyzing signalling pathways and contractile functions using xCelligenceAlthough cardiac function of gene targeted animals can be assessed in vivo through echocardiography analysis, or on the organ level by Langendorff systems, investigating contractile parameters on the cellular level is usually limited by small sample size and high variability: e.g. electrophysiological studies of single cardiomyocytes using patch clamp techniques, measurement of calcium transients in single cells, or force measurements on isolated, skinned cardiac myocytes. In collaboration with Acea we are using the xCELLigence cardio-system to characterize contractile parameters of neonatal mouse cardiomyocyte cultures derived from gene knockout models in a high throughput, high replicate fashion.Using the systemCardiomyocytes of newborn mice were isolated and plated into cardio E-plates. Pilot experiments established optimal culture conditions. You can download a quick protocol here and view the isolation procedure on JOVE.Variables for the optimization process:
Impedance measurementsThe beating behaviour of live cardiomyocytes is recorded label-free in the xCelligence cardio system . Impedance traces are later analysed for several parameters, like beating frequency or amplitude. Because cells are plated in 96-well plates, multiple conditions (cell-density, control vs. knockout cells, addition of small molecule activators/inhibitors) can be tested simultaneously.For more information, go to Acea.
Project outlookWith our partners at Acea, we used the xCelligence system to achieve the following objectives:
Related manuscripts
|
Section 'Sub' Navigation:
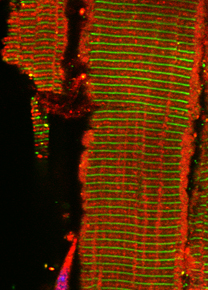
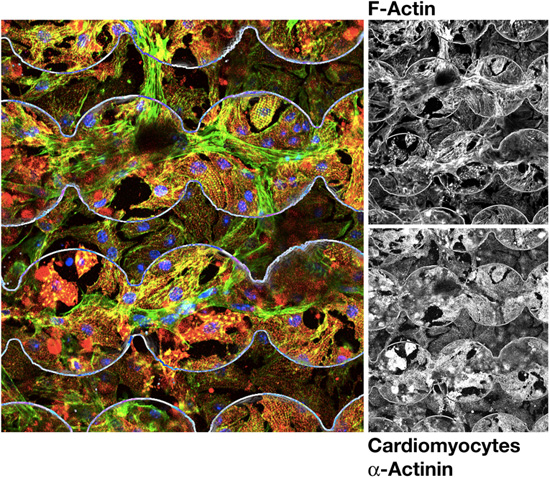 Immunofluorescence image of cardiomyocytes cultured on electrodes to record impedance changes. Fixed cells were stained with antibodies against alpha-actinin (red in overlay, cardiomyocyte marker), filamentous actin (green) and DAPI (blue, nuclei). Electrodes are outlined in white.
Immunofluorescence image of cardiomyocytes cultured on electrodes to record impedance changes. Fixed cells were stained with antibodies against alpha-actinin (red in overlay, cardiomyocyte marker), filamentous actin (green) and DAPI (blue, nuclei). Electrodes are outlined in white.